What makes you YOU? Why are you who you are? What makes you tick and moves your heart? Why are some people gentle and kind where others are hateful and evil? What hurts you? saddens you? Fills you with joy? Why are some people great with children and others don’t care for them at all? What makes one person trust in GOD, and another turn his back?
Everything about you is in your genes!! You carry it with you and pass it on to your children. There are many things in you life that naturally affect your DNA through things you experience and choices you make. But then there are things that change your DNA through methods and madness out of your control. Those who perpetrate these assaults on your being are the folks you idolize, trust and obey. Folks who should be looking out for your best interest.
THE MOST HEINOUS CRIME the Fallen Angels committed was to alter the DNA of God’s creation, transmute the life forms and corrupt all flesh. This was the reason for the flood. ALL FLESH was so badly corrupted that they could not be saved.THIS IS HAPPENING AGAIN IN OUR WORLD TODAY!! WE are reliving the DAYS OF NOAH. The Devil knows his time is short. He is about to unleash all of his power, his creatures, and his weapons upon humanity.
HIS PRIMARY FOCUS is US! He wants to change us completely from the inside out. He is pulling out all the stops and coming after our bodies, minds and spirits. CONTROL… he wants full control. He wants all of us to be his slaves or die.
Ecclesiastes 1:9-10
The thing that hath been, it is that which shall be; and that which is done is that which shall be done: and there is no new thing under the sun. Is there any thing whereof it may be said, See, this is new? it hath been already of old time, which was before us.
spacer
updated: May 16, 2023

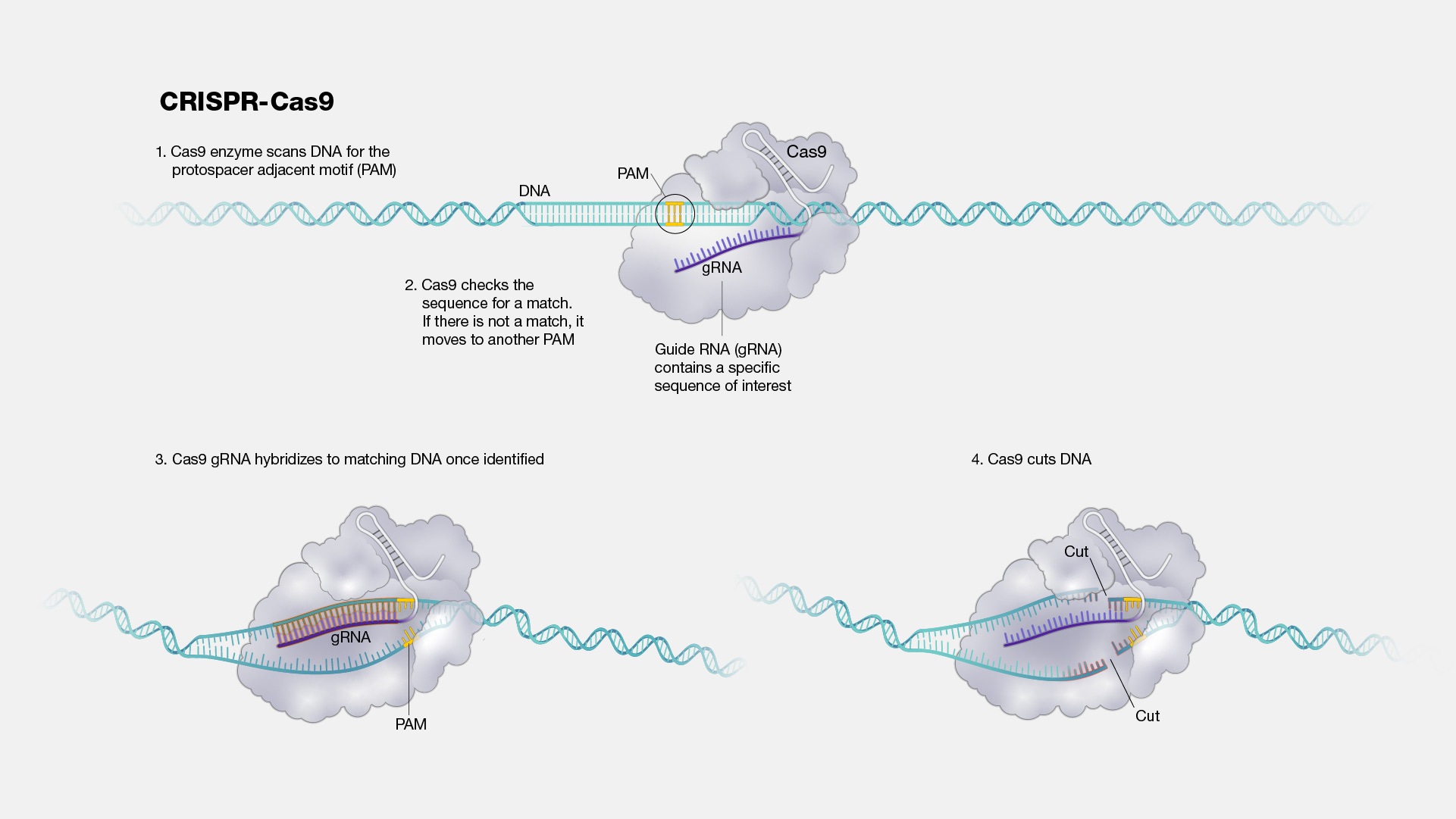
CRISPR (short for “clustered regularly interspaced short palindromic repeats”) is a technology that research scientists use to selectively modify the DNA of living organisms. CRISPR was adapted for use in the laboratory from naturally occurring genome editing systems found in bacteria.
spacer
The Scientific Method vs. Types of Research
Think of the years you’ve spent in science classes and the hundreds of hours you’ve faced teachers drilling you on the “scientific method” (who can forget the poor lima beans squished into dirt-filled paper cups and shoved into dark closets while their siblings enjoyed sunny places on the first-grade window sill?). Do you understand the difference between the “scientific method” and the “types” of research methods?
Think of the scientific method as having four goals (description, prediction, explanation and control). It is important to remember that these goals are the same for anything that can be studied via the scientific method (a chemical compound, a biological organism, or in the case of psychology, behavior). Each goal can be understood in terms of the question that it answers about the entity under investigation. For psychology, each goal answers a particular question with respect to behavior:
- Description: What are the characteristics of the behavior?
- Prediction: How likely is it that the behavior will occur?
- Explanation: What causes the behavior?
- Control: Can I make the behavior happen/ not happen?
spacer
THE LOGICAL STRUCTURE OF THE SCIENTIFIC METHOD
If science is still taught by the apprentice method, it is because the scientific method has never been properly abstracted. Full abstraction discloses that there is only one scientific method. Although its employment in the separate experimental sciences is always mediated by extenuating circumstances, essentially the same set of procedures, conducted in approximately the same order can be discovered in laboratory practices. The scientific method is an on-going process, which nevertheless lends itself to analysis into six well-defined stages. These stages are : observation, hypothesis, experiment, theory, prediction and control. Each of these stages except the first emerges logically from the one before, and each except the last leads logically into the next. Observations are for the purpose of discovering hypotheses; and hypotheses are established in order to test them against fact by experiment, against theory by mathematical calculations, against the application of laws to future particulars by prediction, and finally against the application to practice by the control over fact.
Goals of Science: Describe, Explain, Predict, Control
Scientists have mapped just about everything on earth and gained control over weather, energy, chemicals, minerals, the ocean, etc… NOW, they want to CONTROL YOU. Every aspect of you, including your body, mind and soul/spirit. They are mapping humanity. Our habits, our interests, our personalities, our bodily functions, our thought processes, our emotions and our spirits. Mapping is the LAST STEP before total control. Are you prepared?
spacer
Sequencing 101: Looking Beyond the Single Reference Genome to a pangenome for every species
What is a pangenome?
spacer
Note the familiar symbol of the Trinity. This is a very ancient and esoteric symbol.
spacer
 |
 |
Unless you have an identical twin, no other person has a genome that is identical to yours. The same is true for other animal, plant, and microbial species that reproduce sexually: the genomes of individuals are unique. Less well known, but equally true, is that individual members of a species do not always share even the exact same genes. Nevertheless, scientists mostly use a single reference genome to represent an entire species:
-
-
-
-
-
-
-
- one human genome
- one maize genome
- one Staphylococcus aureus genome
-
-
-
-
-
-
spacer
|
Staphylococcus aureus Infections – Infections – Merck Manuals Consumer … |
The coining of the “pangenome”
Around 2005, geneticists started to explore the concept of the pangenome, originally defined as the entire set of genes possessed by all members of a particular species and then extended to refer to a collection of all the DNA sequences that occur in a species.
It started with bacteria, as many things do. Genomic activity like recombination, mobile genetic elements, and horizontal gene transfer were clearly contributing to individual diversity across the bacterial domain. Some scientists discovered dozens, if not hundreds, of unknown genes when they sequenced new strains.
In 2007, MIT microbiologist Sallie Chisholm (@ChisholmLab_MIT) set out to determine the extent of genetic variation in the marine cyanobacterium Prochlorococcus. Each strain contains approximately 2,000 genes, and Chisholm estimated that a pangenome for Prochlorococcus would be around 6,000 genes, based on an initial set of 12 genome sequences. Eight years later, with 45 strains sequenced, she revised that estimate up to at least 80,000 genes — around four times the number of genes in the human genome — with the core genome for the species comprising only about 1,000 genes, or less than 2 percent of the total gene pool.
“That’s a lot of information shaping that collective,” Chisholm told The Scientist. “[The pangenome view] changes the way you think about what an organism is.”
Why is it important to capture the full range of genetic diversity?
Those looking to create vaccines need to understand the genomic variation and versatility of disease-causing microbes, especially if they are hoping to develop universal vaccines that could provide protection against more than one strain in a species. (Or, if they are looking to create pandemics)
Those studying adaptation to climate change would benefit from a comparison of genes absent or in abundance within species found in different geographic locations and/or environmental conditions. In crop plants, differences in variable genes could have implications on disease resistance, metabolite production, and stress responses.
And with differences in gene number increasingly being associated with disorders including autism, Parkinson’s, and Alzheimer’s diseases, there are strong medical justifications for taking a more variation-centric view of the human species. Variants cannot be identified within regions completely missing from the reference sequence, many of which have been found to be more common than previously thought. (Since they have been studying and experimenting with DNA and with Disease and Immunology, there have been more and more birth defects, chronic diseases, epidemics and vaccine side effects, reactions and deaths. There have also been more algae and aquatic bacteria, as well as more foodborne illnesses. For instance, we had never seen the waters turn blood red before, now it is common, we see it all the time and everywhere.)
What is being done to generate pangenomes?
To answer this question, we sat down with a few scientists to talk about the era of the pangenome and what’s to come.
 |
|
||
| Pangenomes have revealed more genetic diversity within the maize species than between human and chimpanzees. They can and do claim all different animals and plants who share 94% of our genetic makeup… Naturally, lol, we are all made of the earth!! But, God created HUMANS in his own IMAGE and Likeness. No other life form can claim that. | Vesica Pisces – Symbol of Woman’s Vagina, Fertility, Sex. Note the visual of the snake eye, split in the middle. |
spacer
|
meno: 1. a combining form borrowed from Greek, where it meant “month,” used with reference to menstruation in the formation of compound words: menopause. The vagina looks like a mouth.
|
spacer
 16.2K29:41 16.2K29:41MOUNT HERMON 33rd PARALLEL! THE UNITED NATIONS COMPOUND IS LOCATED WHERE THE FALLEN ANGELS “FELL TO EARTH”! WHY? SACRED GEOMETRY? VESICA PISCES! INVERSION! ANCIENT TEMPLE! ALIEN PRESENCE! GUY BRUMMEL : GO FUND ME… You can now FIND ME on GAB TV: NEW AFFILIATE: Sovereign Advisors Additional Affiliates: 2 years ago
 1375:40 1375:40 1336:30 1336:30Old video I made showing one of the many meanings of these symbols, one other is our plane of existance is the vesica pisces and the obelisk is the pole star polaris in the middle 1 year, 3 months ago |
spacer
Original Article continued
spacer
One particularly important crop that has haunted geneticists and breeders for years is maize. It is challenging to sequence because the vast majority of its 2.3 Gb genome, a staggering 85 percent, is made up of highly repetitive transposable elements. Maize is also incredibly diverse in its DNA makeup. As an example, a study comparing genome segments from two inbred lines revealed that half of the sequence and one-third of the gene content was not shared — that’s much more diversity within the species than between humans and chimpanzees, which exhibit around 94 percent sequence similarity.
| Maize is a cultigen; human intervention is required for it to propagate. Whether or not the kernels fall off the cob on their own is a key piece of evidence used in archaeology to distinguish domesticated maize from its naturally-propagating teosinte ancestor.[4] Genetic evidence can also be used to determine when various lineages split. [12 Wikipedia |
| A cultigen (from Latin cultus ‘cultivated’, and gens ‘kind’) or cultivated plant[note 1] is a plant that has been deliberately altered or selected by humans;
Cultigens arise in the following ways:
|
spacer
“The whole notion of a single reference genome for crop plants is an antiquated concept borne out of necessity from the technological limitations of the past. Now with the capability to rapidly generate high-quality references for even the largest crop genomes, we can readily access the full complement of sequence diversity and structural variation within a crop,” says Kevin Fengler, Comparative Genomics Lead at Corteva Agriscience.
So the field was delighted when a collective of 33 scientists released a 26-line maize pangenome reference collection earlier this year. The collection was created using PacBio sequencing, and includes comprehensive, high-quality assemblies of 26 inbreds known as the NAM founder lines. These include the most extensively researched maize lines that represent a broad cross section of modern maize diversity, as well as an additional line containing an abnormal chromosome 10.
It turns out it’s not just maize biology that can be informed by pangenomes. “The high level of diversity in maize is well known, but we see a lot of diversity and structural variation underlying traits of interest in all the crop plants we work on. Creating the first reference genome for a crop genome is a great first step, but things get really interesting as you begin to add more genomes and a more comprehensive view emerges,” adds Fengler.
As for our own species, the current reference genome (GRCh38) — an update of the genome produced by the international Human Genome Project in 2000 and based mostly on DNA from one person — has been added to and annotated through the years, but is still an incomplete sequence and woefully inadequate as a representation of human diversity and genetic variation. Scientists estimate that up to 40 megabases of sequence, including protein-coding regions, are absent from the reference genome.
Several studies using PacBio long reads have reported an average of ~20,000 structural variants (SV) per human genome, most of which fall within repetitive elements and segmental duplications. Furthermore, it does not represent the diploid structure of human genomes. Rather, it is an arbitrary linear combination of different haplotypes, or a mosaic of multiple individuals.
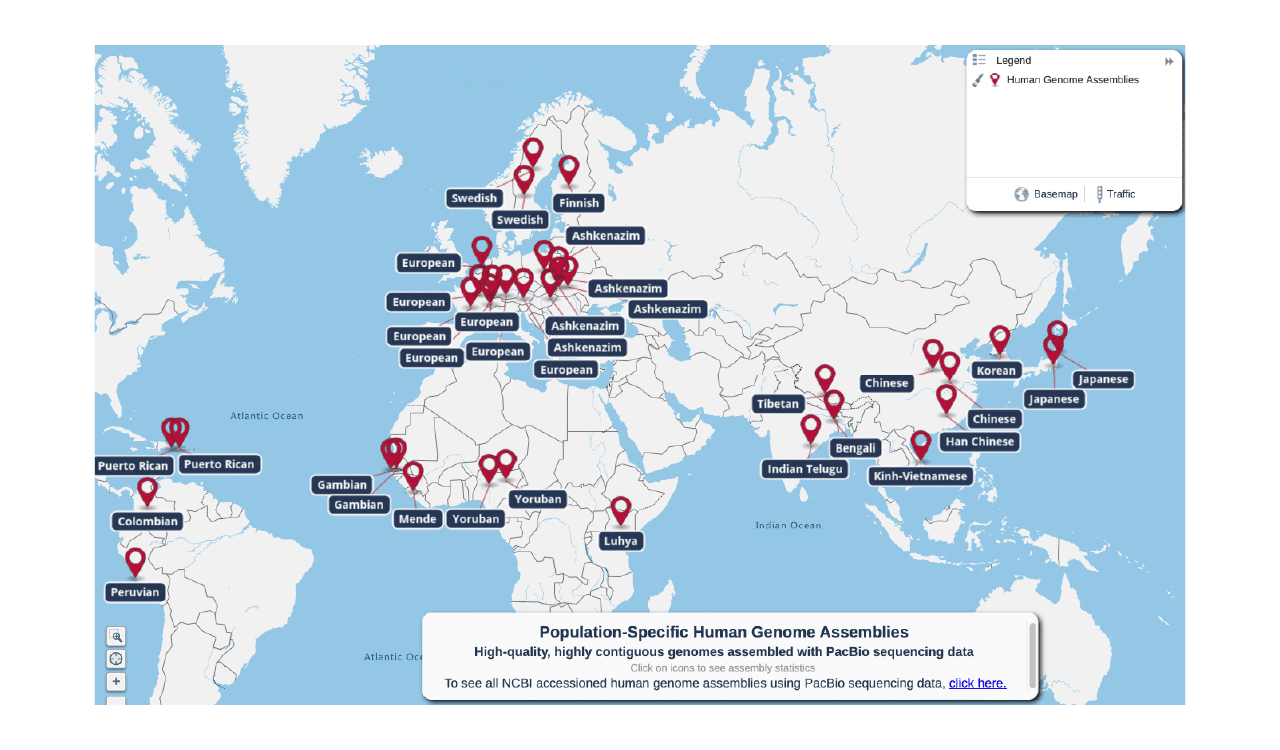
Several groups have undertaken efforts to ensure certain populations are better represented in genomic databases, from Sweden to Tibet to Japan.
When asked the value a pangenome could bring to human research, Fritz Sedlazeck (@sedlazeck), Assistant Professor at Baylor College of Medicine, said, “the pangenome has the potential to represent the diversity of the human population or any species. This eases the re-identification of complex alleles or even haplotypes.”
And it seems the National Human Genome Research Institute agrees, recently committing $30 million towards the creation of a new human pangenome based on high-quality sequencing of 350 individuals from across the human population, to capture all genomic variation observed in human populations.
“One human genome cannot represent all of humanity. The human pangenome reference will be a key step forward for biomedical research and personalized medicine. Not only will we have 350 genomes representing human diversity, they will be vastly higher quality than previous genome sequences,” said David Haussler, director of the University of California Santa Cruz Genomics Institute, which is leading the project.
|
Genome |
How to generate a pangenome
So, what are the most important things to keep in mind when creating a pangenome reference?
First, Fengler says that being able to be confident in your results is really important. “Ideally, all of the references in the pangenome collection will be built with a similar recipe to enable direct comparisons without artifacts from different technologies.” This points to the need for a reliable technology that can be used to generate equivalent quality genomes for many samples with little variability.
Second, the data must be high quality. When asked the importance of long reads to pangenome efforts, Sedlazeck said, “they will be important to distinguish between different alleles/paths in the graph and to characterize novel mutations. Thus, being able to cope with graphs that encode a much higher number of variations to better represent the population.” Along those lines, Fengler adds, “the approach for assembly needs to be robust and accurate such that mis-assembly and sequence errors are not interpreted as structural variation and sequence diversity.”
Lastly, cost and speed have to be taken into account. With the high accuracy of HiFi sequencing, only 10- to 15-fold coverage per haplotype is needed for a high-quality resulting genome assembly, and the analysis time can be cut in half.
“Now researchers no longer need to wait for actionable sequence data,” says Fengler. “For maize, we can generate a high-quality reference genome the same day that the sequencing finishes.”
What’s next in pangenomes?
As pangenome collections grow, scientists have to tackle questions around how to represent a pangenome. “Which variations should be included into a pangenome? Is it all of them? Then you lose specificity in regions. Is it only the common variations? Then you have a problem with disease-causing variations and other complex regions like HLA,” asks Sedlazeck, highlighting the continued work that needs to be done.
In addition, tackling things like annotation, visualization, and relationship management are on Fengler’s mind. “A variety of new pangenome analysis and visualization tools are needed to fully realize the value of having a pangenome collection for each crop.”
And then we have to move into functional and translational analysis. Scientists need to be able to take their newfound understanding of variation at the genome level and see how it impacts phenotypes, and whether the variation can be introduced artificially to influence agronomic traits, for instance.
One thing is for sure: the pangenome era is upon us, and whether you need a pangenome to understand important traits or you build tools to interpret those traits, there will be plenty to work on in the coming years.
Interested in finding out more about HiFi data for pangenome sequencing your organism of interest? Get in touch with a PacBio scientist to scope out your project.
spacer
 I found this image very interesting. A key shaped genome?
I found this image very interesting. A key shaped genome?
KeyGene’s two-part webinar about PanGenomes, Part 2: Constructing and use of a pangenome
PanGenomes describe the available genetic diversity within, for instance, plant species, crops or germplasm collections (or humans) . Because of diminishing costs of genome sequencing, increasing speed of reference genomes construction and advances in bioinformatics, PanGenomes have become an accessible tool. The genome insights delivered by PanGenomes can spectacularly speed up and contribute to plant breeding and research.
KeyGene is well known for its unique combination of extensive expertise in wet lab and in silico research. We offer optimized techniques for high molecular weight plant DNA isolation, the most advanced DNA sequencing platforms and breakthrough bioinformatics pipelines and tools. This combination has already allowed KeyGene to successfully support partners in building and applying plant PanGenomes.
At KeyGene, we underpin the value of PanGenomes and support partners in constructing and using it. Therefore, we organize this two-part webinar on PanGenomes, where we explain what one needs to be able to construct PanGenomes and highlight their value in breeding and research. The two-part webinar will also show KeyGene capabilities in successfully generating and using PanGenomes.
Participation is free of charge.
=> Register here for Part 2 of KeyGene’s two-part webinar on PanGenomes
Part 2: Construction and use of a PanGenome, Thursday 18 March 2021, 16-17h CET
- State-of-the art tools for PanGenome construction at KeyGene, Erwin Datema, Senior Scientist Genome Informatics, KeyGene
- Applications of PanGenome in breeding and research, René Hofstede, Program Scientist, KeyGene
- The value of a PanGenome for crop breeding, Allen Sessions, Senior Principle Scientist and Crop Genetics Team Lead, BASF
 Dr Allen Sessions holds a PhD in Plant Biology from the University of California, Berkeley. He has held leading positions at Syngenta and Bayer CropScience working on genotyping, marker-trait associations and next-generation sequencing applications in cotton, maize, and soybean. Currently, Dr Sessions is a senior principal scientist and Group Leader of Crop Genetics at BASF where his team focuses on trait discovery and deployment for cotton and soybean breeding while applying various NGS technologies.
Dr Allen Sessions holds a PhD in Plant Biology from the University of California, Berkeley. He has held leading positions at Syngenta and Bayer CropScience working on genotyping, marker-trait associations and next-generation sequencing applications in cotton, maize, and soybean. Currently, Dr Sessions is a senior principal scientist and Group Leader of Crop Genetics at BASF where his team focuses on trait discovery and deployment for cotton and soybean breeding while applying various NGS technologies.
=> Register now for Part 2 of KeyGene’s two-part webinar on PanGenomes
Throughout the world’s existence the fallen Angels and those who serve them have worked very hard to convince humanity that they can thrive without the intervention of the Creator. They have had thousands of years to erase the truth, and fill the world with deception. They have been very successful as most of the world today does not even believe that God exists, or that the Devils exist, or that judgement is coming.
There are many pagan religions once again in our world. Too many to go into here, but all religions really fall somewhere under the following:
Pantheism is the belief that God and the universe are one and the same. There is no dividing line between the two. Pantheists believe that everything is part of an all-encompassing, immanent God. All forms of reality may then be considered either modes of that Being, or identical with it. They view God as immanent and impersonal. This belief system grew out of the Scientific Revolution, and pantheists generally are strong supporters of scientific inquiry, as well as religious toleration. Pantheists don’t believe there is a God that made the earth or defined gravity, but, rather, God is the earth and gravity and everything else in the universe. Because God is uncreated and infinite, the universe is likewise uncreated and infinite and ever changing.. God did not choose one day to make the universe. Rather, it exists precisely because God exists, since the two are the same thing. Because all things are ultimately God, all approaches to God can conceivably lead to an understanding of God. Each person should be allowed to pursue such knowledge as they wish. This does not mean, however, that pantheists believe every approach is correct. They generally do not believe in an afterlife, for example, nor do they find merit in strict dogma and ritual. Pantheist thought can be found within animistic beliefs and tribal religions throughout the world as an expression of unity with the divine, specifically in beliefs that have no central polytheist or monotheist personas. Pantheism derives from the Greek word πᾶν pan (meaning “all, of everything”) and θεός theos (meaning “god, divine”).
Animism (from Latin: anima meaning ‘breath, spirit, life‘) is the belief that objects, places, and creatures all possess a distinct spiritual essence. Animism perceives all things—animals, plants, rocks, rivers, weather systems, human handiwork, and in some cases words—as animated and alive. Animism is used as a term for the belief system of many Indigenous peoples. Although each culture has its own mythologies and rituals, animism is said to describe the most common, foundational thread of indigenous peoples’ “spiritual” or “supernatural” perspectives. Animists believe that all material phenomena have agency, that there exists no categorical distinction between the spiritual and physical world, and that soul, spirit, or sentience exists not only in humans but also in other animals, plants, rocks, geographic features (such as mountains and rivers), and other entities of the natural environment. Examples include water sprites, vegetation deities, and tree spirits, among others. Animism may further attribute a life force to abstract concepts such as words, true names, or metaphors in mythology. The origin of the word comes from the Latin word anima, which means life or soul.
Polytheism is the belief in many divine beings, who typically have to beworshippedor, if malevolent, warded off with appropriate rituals. Like some other religions Polytheism identifies natural forces and objects as divinities. classified in three categories celestial, atmospheric, and earthly. In pastoral times when people survived through hunting and agriculture the forces of nature were vital to survival. Ancient peoples developed religions that exhibit rites connected with fertility. The sun’s vitality was seen in the cyclical effects of causing things to grow and wither. Because of its dominance over the world, the sun was often seen as all-knowing, and thus sky gods of various cultures were seen as highly powerful and knowledgeable. The sky was associated with creation in contrast the moon was rarely seen as important (though in Ur, a city of ancient southern Babylonia, the moon god Sin was supreme). The role of the sky god in ensuring food and in providing light and warmth, over against the chaotic effects of darkness, was a theme of various myths of the cosmic drama and was one main reason for the connection in mythic thought between creation and light. Important in the development of fertility religion were the “dying and rising” gods, such as Adonis, Attis, Osiris, and Tammuz. Their cults had a new life in the mystery cults of the Greco-Roman world, where the original agricultural significance of the rites was transformed into more personal and psychological terms. On earth, besides the divine mother out of whose womb plant life has its birth, there are a host of divinities connected with agricultural and pastoral life. In addition, sacred significance is often attached to features of the particular environment in which a given group finds itself. Thus, sacred mountains, such as Olympus in Greece, have their resident deities, and a river, such as the Ganges (Ganga), may be divinized. Underground rivers have special significance in connecting with the underworld, or nether regions, which can be important as the place of repose of the dead but also as the matrix for the re-creation of life. Geographical locations can also have cosmic significance; e.g., Delphi, Greece, was known as the navel of the earth. Further, many cultures have gods and goddesses associated with the sea.
Scientists, Technologists, Scholars and the Wealthy Elite want you to believe that they do not believe in GOD or are not religious at all…they are developed beyond such nonsense. That is a flat out lie. They serve their gods and goddesses faithfully, and don’t make a move without them. This is clearly demonstrated in their symbolism and numerology.
The TRUTH is that there is ONLY ONE TRUE and LIVING GOD who CREATED ALL THINGS. Those who know him enjoy the peace that passes all understanding. They are comforted in knowing that there is no lack with GOD and that He is always in control.
God created us in His image. That means that our bodies were designed to be like His. Everything about our bodies was designed to function properly, to be self healing, to work together as single whole.
The elite do all that they can to reduce us to nothing more than a lump of cells thrown together by accident and with no more importance that a stone or plant or spec of dust. DON’T BUY THEIR LIES.
Here you will see that the have terms for the precious gift of new life created by GOD that reduce it to insignificant, disposable tissue.

| Dictionary entry | Language | Definition |
|---|---|---|
| *jungō | Proto-Italic (itc-pro) | To join. To yoke. |
| *yung- | Proto-Indo-European (ine-pro) | To Join |
| iungere | Latin (lat) | |
| *yugóm | Proto-Indo-European (ine-pro) | Yoke. |
| *dzugón | Proto-Hellenic (grk-pro) | |
| *dzugós | Proto-Hellenic (grk-pro) | |
| ζυγός | Ancient Greek (grc) | joined/yoked |
| ζυγόω | Ancient Greek (grc) | |
| ζυγωτός | Ancient Greek (grc) | |
| zygote | English (eng) | A fertilized egg cell. |
Zygote – Wikipedia
|
Etymology: Came from the Greek word “gametes” meaning “husband” and “gamete” meaning “wife.” … Zygote formed by the process of fertilization undergoes rapid mitotic divisions to form blastocyst and subsequently the fetus. Morphology. The sperm is a tadpole-like structure that has a head, a mid section and a tail.
|
|
gamete – găm′ēt″, gə-mēt′ – noun
|
|
zygospore – zī′gə-spôr″, zĭg′ə- noun
|
|
An oospore is a thick-walled sexual spore that develops from a fertilized oosphere in some algae, fungi, and oomycetes. They are believed to have evolved either through the fusion of two species or the chemically-induced stimulation of mycelia, leading to oospore formation. Wikipedia
|
Human Pangenome Reference Consortium
We have released a new high-quality collection of reference human genome sequences that includes genome sequences of 47 people, with the the goal of increasing that number to 350 by mid-2024.
Link to Publications:
A draft human pangenome reference;
Increased mutation rate and gene conversion within human segmental duplications;
Recombination between heterologous human acrocentric chromosomes;
Pangenome graph construction from genome alignment with minigraph-cactus.
We Need More Than One Human Reference Genome
The HPRC was started to help migrate common genomic analysis to use a pangenome so diverse populations are better represented.

What are we doing?
Milestones
|
Year 1
47
|
Current Production
100+
Next Round
|
Completion
350+
Usable Mature
|
Collaboration Map

spacer
|
phylogeny – fī-lŏj′ə-nē – noun
|
|
Taxonomy Definition & Meaning – Merriam-Webster |
spacer
Goals of the Tree of Life
The Tree of Life Web Project is an online database that compiles information about biodiversity and the evolutionary relationships of all organisms. Content for the project is compiled collaboratively by hundreds of biologists and amateur contributors from all over the world. The project is non-profit, funded by the U. S. National Science Foundation and the University of Arizona. All of our services are free and open to the public.
We envisage the Tree of Life being used by people interested in locating information about a particular group of organisms, by biologists seeking identification keys, figures, phylogenetic trees, and other systematic information for a group of organisms, and by educators teaching about organismal diversity. The ToL project was originally designed for biologists. However, given the response of other people to the project, including middle and high school students, we are encouraging authors to include information of interest to non-biologists, and we now also accept contributions from students and other amateur scientists and nature lovers. For more information, please see Contributing Images and Other Media to the Tree of Life and Learning with the Tree of Life.
The basic goals of the Tree of Life project are:
- To present information about every species and significant group of organisms on Earth, living and extinct, authored by experts in each group.
- To present a modern scientific view of the evolutionary tree that unites all organisms on Earth.
- To aid learning about and appreciation of biological diversity and the evolutionary Tree of Life.
- To share information with other databases and analytical tools, and to phylogenetically link information from other databases.
 spacer
spacerAbout the ToL Navigation Picture
We’ve been getting a lot of inquiries about the tree of life picture on our home page. It is important to note that the major function of this picture is to help visitors to the ToL web site to quickly navigate to pages of some of the major groups of organisms. In order to serve this purpose, we had to use a greatly simplified representation of the tree of life.

|
 Tree of life The tree of life is a fundamental archetype in many of the world’s mythological, religious, and philosophical traditions. It is closely related to the concept of the sacred tree. The concept of the tree of life may have originated in Central Asia, and absorbed by other cultures, such as Scandinavian mythology and Altai shamanism.
Wikipedia |
What Is the Tree of Life?
The tree of life, first referenced in the book of Genesis, is a life-giving tree created to support the physical and spiritual life of mankind. The tree was established by God in the Garden of Eden: “And out of the ground the LORD God made to spring up every tree that is pleasant to the sight and good for food. The tree of life was in the midst of the garden, and the tree of the knowledge of good and evil.” (Genesis 2:9).
In his transgression, Adam lost his everlasting life. The tree of life in Eden must have played a role in sustaining the life of Adam and Eve. Therefore, Adam would have “lived forever,” even in his fallen state, if he had eaten the tree of life after his sin.
Then the LORD God said, “Behold, the man has become like one of us in knowing good and evil. Now, lest he reach out his hand and take also of the tree of life and eat, and live forever–” therefore the LORD God sent him out from the garden of Eden to work the ground from which he was taken. He drove out the man, and at the east of the garden of Eden he placed the cherubim and a flaming sword that turned every way to guard the way to the tree of life.
spacer
What is the tree of life?
The tree of life shows how all life on earth is related. Each leaf represents a different species. The branches show how these many species evolved from common ancestors over billions of years. In our interactive tree of life you can explore the relationships between 2,235,076 species and wonder at 105,409 images on a single zoomable page.

Eliphas Levi declared that by arranging the Tarot cards according to a definite order man could discover all that is knowable concerning his God, his universe, and himself. When the ten numbers which pertain to the globes (Sephiroth) are combined with the 22 letters relating to the channels, the resultant sum is 32–the number peculiar to the Qabbalistic Paths of Wisdom. These Paths, occasionally referred to as the 32 teeth in the mouth of the Vast Countenance or as the 32 nerves that branch out from the Divine Brain, are analogous to the first 32 degrees of Freemasonry, which elevate the candidate to the dignity of a Prince of the Royal Secret. Qabbalists also consider it extremely significant that in the original Hebrew Scriptures the name of God should occur 32 times in the first chapter of Genesis.
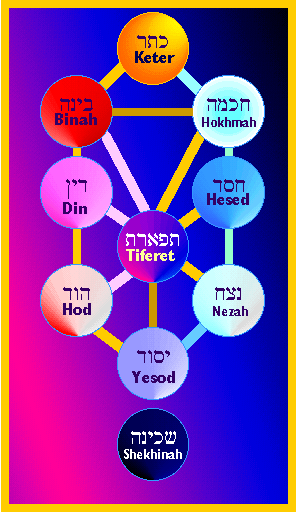 The Jewish mystical doctrine known as “Kabbalah” (=”Tradition”) is distinguished by its theory of ten creative forces that intervene between the infinite, unknowable God (“Ein Sof”) and our created world.
The Jewish mystical doctrine known as “Kabbalah” (=”Tradition”) is distinguished by its theory of ten creative forces that intervene between the infinite, unknowable God (“Ein Sof”) and our created world.
(according to Kabbalah it is) Through these powers God created and rules the universe, and it is by influencing them that humans cause God to send to Earth forces of compassion or severe judgment.
The right side represents the principles of unity, harmony and benevolence, as embodied in theSefirah of Hesed. It is associated with the bestowing of generous goodness upon our world. It is the masculine side.
The Left side of the Sefirot structure is the side of power and strict justice, the values embodied in the Sefirah of Din. It is the female side, representing the fearsome awe of God, and the principles of separation and distinction. The unrestrained dominion of the Left side gives rise to Evil.
The Middle column of the Sefirot structure represents the ideal balance of divine mercy and justice. This harmony is best represented in the Sefirah of Tiferet, which exists midway between the extremes of Din and Hesed, female and male. Following traditional Rabbinic perception, the Kabbalah recognizes that the universe could not survive if it were founded only upon justice or only upon mercy.
You can learn about the individual Sefirot by clicking on their pictures in the diagram above or on the text below:
The TEN SEFIROT/ FORCES / ANGELIC/DEMONIC BEINGS
| keter ʿelyon (“supreme crown”) ḥalhma (“wisdom”) bina (“intelligence”) ḥesed (“love”) gevura (“might”) tif ʾeret (“beauty”) netzah (“eternity”) hod (“majesty”) yesod (“foundation”) malkhut (“kingship”). Source |
spacer
sefirot, also spelled sephiroth, singular sefira or sephira, in the speculations of esoteric Jewish mysticism (Kabbala), the 10 emanations, or powers, by which God the Creator was said to become manifest. The concept first appeared in the Sefer Yetzira (“Book of Creation”), as the 10 ideal numbers.
In the development of Kabbalistic literature, the idea was expanded and elaborated to denote the 10 stages of emanation from En Sof (the Infinite; the unknowable God), by which God the Creator can be discerned. Each sefira refers to an aspect of God as Creator; the rhythm by which one sefira unfolds to another was believed to represent the rhythm of creation. The mystical nature of the sefirot and the precise way in which they function were often disputed. Kabbalists used them as one of their principal subjects of mystical contemplation, despite vigorous criticism that such speculations were implicitly heretical.
Ein Sof
The Ein Sof (lit: without end) is an important concept in Jewish Kabbalah. Generally translated as “infinity” and “endless”, the Ein Sof represents the formless state of the universe before the self-materialization of God. In other words, the Ein Sof is God before he decided to become God as we now know him.[4]
The sefirot are divine emanations that come from the Ein Sof in a manner often described as a flame. The sefirot emanate from above to below. As the first Sefira is closest to Ein Sof, it is the least comprehensible to the human mind, while in turn the last is the best understood because it is closest to the material world that humanity dwells on.[4]
All the sephirot are likened to different parts of the body and the tree itself to an homunculus. Wikipedia
spacer
If you think that UFOs and Aliens are something new…guess again. This same lie/deception has been perpetrated on humanity off and on throughout history. This comes from the Fallen Angels who want you to believe that they are gods from space, returning to earth to save humanity. DON’T BUY THE LIE!
The name Aramatena is the 12th Lyran Stargate and has been used to describe schematics of the future earth blueprint in its crystalline form in the Avatar matrix Universe. The Aurora or Ascension Earth is located in the Andromedan Time Matrix which is found through the passage that interfaces between the Milky Way and Andromeda Galaxy, and that is connected through the networks of the Polarian Gate hosting system.
Our planet Earth exists in three main formed identities in this Universal Time Matrix. 3D Earth is called Earth or Terra, 5D Earth is Called Tara and 7D earth is called Gaia. Different E.T. Races and extra dimensionals may call these future Earths by different names based on their language. (Such as Urantia) They are all planet Earth at different stages of evolution in the Timelines.

They are REBUILDING us. THE BUILDERS. They are forming us in their image with demonic spirits ruling over us from the inside. They are corrupting our flesh in every way possible.
Practical Graphical Pangenomics
Parts of the pangenome
In the pangenome, we can identify three sets of genes: Core, Shell, and Cloud genome. The Core genome comprises the genes that are present in all genomes analyzed. To avoid dismissing families due to sequencing artifacts some authors consider the softcore (>95% occurrence). The Shell genome consists of the genes shared by the majority of genomes (10-95% occurrence). The gene families present in only one genome or <10% occurrence are described as Dispensable or Cloud genome.
Core
Is the part of the pangenome that is shared by every genome in the tested set. Some authors have divided the core pangenome in hard core, those families of homologous genes that has at least one copy of the family shared by every genome (100% of genomes) and the soft core or extended core,[15] those families distributed above a certain threshold (90%). In a study that involves the pangenomes of Bacillus cereus and Staphylococcus aureus, some of them isolated from the international space station, the thresholds used for segmenting the pangenomes were as follows: “Cloud,” “Shell,” and “Core” corresponding to gene families with presence in <10%, 10 to 95%, and >95% of the genomes, respectively.[16]
The core genome size and proportion to the pangenome depends on several factors, but it is especially dependent on the phylogenetic similarity of the considered genomes. For example, the core of two identical genomes would also be the complete pangenome. The core of a genus will always be smaller than the core genome of a species. Genes that belong to the core genome are often related to house keeping functions and primary metabolism of the lineage, nevertheless, the core gene can also contain some genes that differentiate the species from other species of the genus, i.e. that may be related pathogenicity to niche adaptation.[17]
Shell
Is the part of the pangenome shared by the majority of the genomes in a pangenome.[18] There is not a universally accepted threshold to define the shell genome, some authors consider a gene family as part of the shell pangenome if it shared by more than 50% of the genomes in the pangenome.[19] A family can be part of the shell by several evolutive dynamics, for example by gene loss in a lineage where it was previously part of the core genome, such is the case of enzymes in the tryptophan operon in Actinomyces,[20] or by gene gain and fixation of a gene family that was previously part of the dispensable genome such is the case of trpF gene in several Corynebacterium species.[21]
Cloud
The cloud genome consists of those gene families shared by a minimal subset of the genomes in the pangenome,[22] it includes singletons or genes present in only one of the genomes. It is also known as the peripheral genome. Gene families in this category are often related to ecological adaptation.[citation needed]

GENOMICS
Roary: Analysis of Prokaryote Pan Genome on a large-scale
The Microbial Pan Genome is the union of genes shared by genomes of interest. This term was first used by Medini in 2005.
Since then, microbial genome data has been enormously increased, so to study processes such as selection and evolution, the construction of pan genome of species is required. But construction of pan genome from the real data available is very difficult and would not be accurate due to fragmented assemblies, poor annotation and also the contamination,i.e., microbial organisms can rapidly acquire genes from other organisms. Therefore, Andrew J. Page et al have developed
a new method to generate the pan genome of a set of related prokaryotic isolates and named the tool as ‘Roary’. It deals with thousands of isolates in a feasible time.
How Roary Works?
One annotated assembly per sample is input in the Roary from which coding regions are extracted and converted in to protein sequences, and all the partial sequences are removed and pre clustered using CD-HIT (a fast program for clustering and comparing). This produces a reduced set of protein sequences. These reduced sequences are compared all-against-all with the help of BLASTP with a user defined percentage sequence identity (default 95%). Now, by using conserved neighborhood genes, homologous groups are split in to true orthologs. Finally, a graph is constructed showing the relationships of the clusters based on the order of occurrence in the input sequences.
|
Sequence homology is the biological homology between DNA, RNA, or protein sequences, defined in terms of shared ancestry in the evolutionary history of life.Two segments of DNA can have shared ancestry because of three phenomena: either a speciation event (orthologs), or a duplication event (paralogs), or else a horizontal (or lateral) gene transfer event (xenologs).
|

Fig.1 Effect of dataset size on the wall time of multiple applications.
That’s how the orthologous genes of prokaryotes can be easily identified and the microbial evolution can be well studied. It is done on a large scale covering a large data set to analyse the pan genomes of prokaryotes. Other tools have also been made earlier than Roary for the same purpose,namely, PanOCT and PGAP, but Roary is more fast, heuristic and most feasible tool among them.
spacer
|
Key points: Prokaryotesare single-celled organisms belonging to the domains Bacteria and Archaea. Prokaryotic cells are much smaller than eukaryotic cells, have no nucleus, and lack organelles. All prokaryotic cells are encased by a cell wall. Many also have a capsule or slime layer made of polysaccharide.
|
spacer

Building a beautiful Pangenome
Cool tech and ingenuity are building a beautiful Pangenome. It’s a reference genome representing the full spectrum of human genetic variation.
For twenty years, geneticists at the Human Genome Project have built the Reference Genome to give “the world a resource of detailed information about the structure, organization, and function of the complete set of human genes.”
Looking into the nano-spaces of our genome to get precious information has been tough because DNA is bundled tightly into chromosomes and packed into the tiny nucleus in the cell. Especially difficult to unravel are the telomeres and centromeres.
| What Are Telomeres? Telomeres are segments of DNA at the end of our chromosomes. Scientists frequently compare them to the plastic tips of shoelaces that keep the laces together. (1) Telomeres function similarly, preventing chromosomes from fraying or tangling with one another. When that happens, it can cause genetic information to get mixed up or destroyed, leading to cell malfunction, increasing the risk of disease or even shortening lifespans.Each time a cell divides, its telomeres become shorter. After years of splicing and dicing, telomeres become too short for more divisions. At this point, cells are unable to divide further and become inactive, die or continue dividing anyway — an abnormal process that’s potentially dangerous. |
|
Centromere – Definition, Function and Types | Biology Dictionary |
Telomeres cap the end of our chromosomes. Centromeres are the knotted-looking section in the middle. Both have thousands of tightly wrapped, repeated segments of DNA and hold essential information.

During the past two decades, scientists used NexGen technology to “see” and “read” the exact ATCG sequence on the chromosomes. But Nexgen is a short-read method with fundamental limitations. There are gaps in the sequence data. It can’t make sense of the tightly wrapped telomeres and centromeres, nor can it fully sequence non-coding regions.
With long-read tech, scientists can “see every crater, every color, from something that only had the blurriest understanding of before.” And make thrilling discoveries, “researchers have uncovered more than 100 new genes that may be functional and have identified millions of genetic variations between people. Some of those differences probably play a role in diseases.”
Advanced sequencing technology also plays a role in addressing the diversity problem. The current Reference Genome DNA is from mostly European populations. It lacks the full spectrum of worldwide genetic variation. So scientists are building a Human Pangenome with long-read tech. They’re accumulating so much data that two projects are set up to sequence, map, and analyze it: (ALWAYS REMEMBER MAPPING IS THE LAST STEP TO CONTROL)
Telomere to Telomere Project fills the gaps by sequencing chromosomes end-to-end from people worldwide with diverse ancestry. They’ve already identified “hundreds of thousands of novel variants per sample — a new frontier for evolutionary and biomedical discovery.” More about the T2T project here.
Human Genome Reference Program will gather “high-quality gapless sequences from ancestrally diverse people.” It will consolidate data from many sources, such as the T2T project. Other researchers are improving sequence technology, and bioinformatics scientists are designing tools for better data analysis. More about the program here.
The Human Pangenome Reference Consortium “aims to issue a new pangenome reference assembly to the international community that reflects the full range of genomic diversity across the globe. We are committed to achieving this goal in a responsible and ethical manner, with explicit attention to community engagement, inclusion, and fair representation.” More about the consortium here.
Put them together, and voila! Pulling data from a wide variety of sources will take years to sort and analyze. The result won’t be just one Reference Genome. What started more than twenty years ago with “the one” will expand into a worldwide compilation of hundreds of data sets — beautiful Pangenomes for a unified representation of the human species.
Finding wonder and beauty in the science of us. From an artistic perspective, we could say scientists are gathering all the subtle hues of human variation. And we are mining the new data for hidden gems. What can we imagine with the full spectrum of human genetic diversity? What would you write, draw, compose, sing about your genome?
One thing is for sure: the pangenome era is upon us, and whether you need a pangenome to understand important traits or you build tools to interpret those traits, there will be plenty to work on in the coming years.
Thomas Lin Pedersen @thomasp85 · Super excited to team up with a Danish design juggernaut with art, sound, and visuals in their DNA and help them enter web3. Along with fellow artists @shavonnewong_ and @Hackatao Quote Tweet Bang & Olufsen @BangOlufsen · It’s official. The #DNACollection is here. Read more about it below: on.beo.com/dnacollection
|
roseens (Danish)Noun – roseens (common)
This is the meaning of rose: rose (Danish) Origin & history IFrom late Old Norse rós, rósa, from Middle Low German rōse, from Latin rosa (“rose”). Pronunciation IPA: /roːsə/ Noun rose
Origin & history II From French rosé.Alternative forms roséPronunciation IPA: /rose/ Noun rose
Origin & history IIIFrom Old Norse hrósa, whence dialectal English roose. Pronunciation IPA: /roːsə/ Verb |
|
Etymology 1 . From Old Norse kelda, from Proto-Germanic *kaldijǭ, cognate with Norwegian kjelde, Swedish källa. Derived from *kaldaz (” kold “). Noun . kilde c (singular definite kilden, plural indefinite kilder) spring, fountain, source (a place where water emerges from the ground)
|
·Since 2003, all humans on Earth have been compared to only one reference genome. We need reference genomes for every ancestry – a pangenome.


spacer
The microbial pangenome is the union of genes shared by genomes of interest. The core genome is the intersection of genes of these genomes, thus core genes are genes present in all strains. The accessory genome (also: variable, flexible, dispensable genome) refers to genes not present in all strains of a species. These include genes present in two or more strains or even genes unique to a single strain. Acquired antibiotic resistance genes are typically genes of the accessory genome.
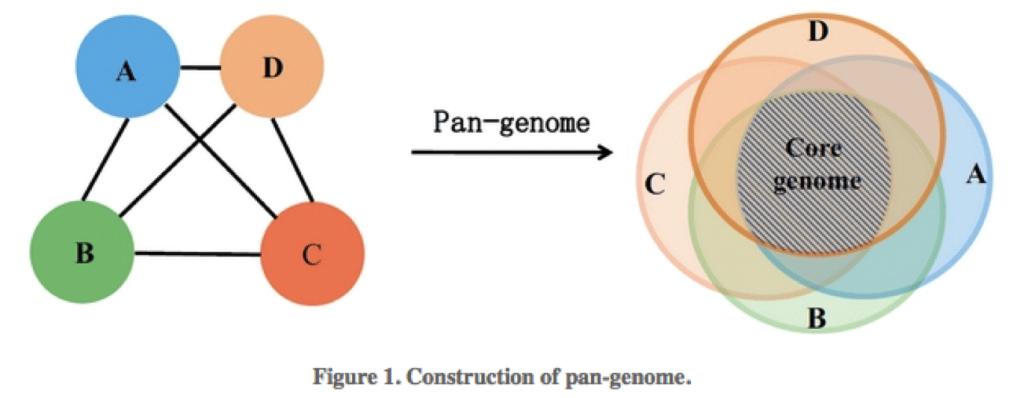
Pangenome analysis
Comparing every gene of every genome to every other gene in the dataset is an enourmous task, and takes a long time even if automated. Roary is a pipeline to determine genes of the core and pangenome. It takes a few short-cuts such as clustering instead of pair-wise alignment and can perform this task in a relatively short time frame. An excellent step-by-step tutorial can be found here
Visualization
Some genes are present in all genomes, some are present in some and absent in others. Data on presence and absence of genes was collected in a matrix called gene_presence_absence.csv. Clustering of this information was used to build a tree (available as accessory_binary_genes.fa.newick). As a next step we are going to visualize this clustering.
Key Points
- The microbial pangenome is the union of genes in genomes of interest.
- The microbial core genome is the intersection of genes shared by genomes of interest.
- Roary is a pipeline to determine genes of the pangenome.

Scientists have published the first human “pangenome” — a full genetic sequence that incorporates genomes from not just one individual, but 47.
These 47 individuals hail from around the globe and thus vastly increase the diversity of the genomes represented in the sequence, compared to the previous full human genome sequence that scientists use as their reference for study. The first human genome sequence was released with some gaps in 2003 and only made “gapless” in 2022. If that first human genome is a simple linear string of genetic code, the new pangenome is a series of branching paths.
The ultimate goal of the Human Pangenome Reference Consortium, which published the first draft of the pangenome on Wednesday (May 10) in the journal Nature, is to sequence at least 350 individuals from different populations around the world. Although 99.9% of the genome is the same from person to person, there is a lot of diversity found in that final 0.1%.
“Rather than using a single genome sequence as our coordinate system, we should instead have a representation that is based on the genomes of many different people so we can better capture genetic diversity in humans,” Melissa Gymrek, a genetics researcher at the University of California, San Diego, who was not involved in the project, told Live Science.
Related: More than 150 ‘made-from-scratch’ genes are in the human genome. 2 are totally unique to us.

A reference for health
The first full human genome sequence was completed in 2003 by the Human Genome Project and was based on one person’s DNA. Later, bits and pieces from about 20 other individuals were added, but 70% of the sequence scientists use to benchmark genetic variation still comes from a single person.
Geneticists use the reference genome as a guide when sequencing pieces of people’s genetic codes, Arya Massarat, a doctoral student in Gymrek’s lab who co-authored an editorial about the new research with her in the journal Nature, told Live Science. They match the newly decoded DNA snippets to the reference to figure out how they fit within the genome as a whole. They also use the reference genome as a standard to pinpoint genetic variations — different versions of genes that diverge from the reference — that might be linked with health conditions.
But with a single reference mostly from one person, scientists have only a limited window of genetic diversity to study.
The first pangenome draft now doubles the number of large genome variants, known as structural variants, that scientists can detect, bringing them up to 18,000. These are places in the genome where large chunks have been deleted, inserted or rearranged. The new draft also adds 119 million new base pairs, meaning the paired “letters” that make up the DNA sequence, and 1,115 new gene duplication mutations to the previous version of the human genome.
“It really is understanding and cataloging these differences between genomes that allow us to understand how cells operate and their biology and how they function, as well as understanding genetic differences and how they contribute to understanding human disease,” study co-author Karen Miga, a geneticist at the University of California, Santa Cruz, said at a press conference held May 9.
The pangenome could help scientists get a better grasp of complex conditions in which genes play an influential role, such as autism, schizophrenia, immune disorders and coronary heart disease, researchers involved with the study said at the press conference.
For example, the Lipoprotein A gene is known to be one of the biggest risk factors for coronary heart disease in African Americans, but the specific genetic changes involved are complex and poorly understood, study co-author Evan Eichler, a genomics researcher at the University of Washington in Seattle, told reporters. With the pangenome, researchers can now more thoroughly compare the variation in people with heart disease and without, and this could help clarify individuals’ risk of heart disease based on what variants of the gene they carry.
spacer
Related: As little as 1.5% of our genome is ‘uniquely human’
A diverse understanding
The current pangenome draft used data from participants in the 1000 Genomes Project, which was the first attempt to sequence genomes from a large number of people from around the world. The included participants had agreed for their genetic sequences to be anonymized and included in publicly available databases.
The new study also used advanced sequencing technology called “long-read sequencing,” as opposed to the short-read sequencing that came before. Short-read sequencing is what happens when you send your DNA to a company like 23andMe, Eichler said. Researchers read out small segments of DNA and then stitch them together into a whole. This kind of sequencing can capture a decent amount of genetic variation, but there can be poor overlap between each DNA fragment. Long-read sequencing, on the other hand, captures big segments of DNA all at once.
RELATED STORIES
—Humans’ big-brain genes may have come from ‘junk DNA’
—Rosalind Franklin knew DNA was a helix before Watson and Crick, unpublished material reveals
—Smallest genome of living creature discovered
While it’s possible to sequence a genome with short-read sequencing for about $500, long-read sequencing is still expensive, costing about $10,000 a genome, Eichler said. The price is coming down, however, and the pangenome team hopes to sequence their next batches of genomes at half that cost or less.
The researchers are working to recruit new participants to continue to fill in diversity gaps in the pangenome, study co-author Eimear Kenny, a professor of medicine and genetics at the Institute for Genomic Health at Icahn School of Medicine at Mount Sinai in New York City, told reporters. Because genetic information is sensitive and because different rules govern data-sharing and privacy in different countries, this is delicate work. Issues include privacy, informed consent, and the possibility of discrimination based on genetic information, Kenny said.
Already, researchers are uncovering new genetic processes with the draft pangenome. In two papers published in Nature alongside the work, researchers looked at highly repetitive segments of the genome. These segments have traditionally been difficult to study, biochemist Brian McStay of the National University of Ireland Galway, told Live Science, because sequencing them via short-read technology makes it hard to understand how they fit together. The long read technology allows for long chunks of these repetitive sequences to be read at once.
The studies found that in one type of repetitive sequence, known as segmental duplications, there is a larger than expected amount of variation, potentially a mechanism for the long-term evolution of new functions for genes. In another type of repetitive sequence that is responsible for building the cellular machines that create new proteins, though, the genome stays remarkably stable. The pangenome allowed researchers to discover a potential mechanism for how these key segments of DNA stay consistent over time.
“This is just the start,” McStay said. “There will be a whole lot of new biology that will come out of this.“
Citations
Clostridium baratii strains are rare opportunistic pathogens associated with botulism intoxication. They have been isolated from foods, soil and be carried asymptomatically or cause botulism outbreaks. Is not taxonomically related to Clostridium botulinum, but some strains are equipped with BoNT/F7 cluster. Despite their relationship with diseases, our knowledge regarding the genomic features and phylogenetic characteristics is limited. We analyzed the pangenome of C. baratii to understand the diversity and genomic features of this species. We compared existing genomes in public databases, metagenomes, and one newly sequenced strain isolated from an asymptomatic subject. The pangenome was open, indicating it comprises genetically diverse organisms. The core genome contained 28.49% of the total genes of the pangenome. Profiling virulence factors confirmed the presence of phospholipase C in some strains, a toxin capable of disrupting eukaryotic cell membranes. Furthermore, the genomic analysis indicated significant horizontal gene transfer (HGT) events as defined by the presence of prophage genomes. Seven strains were equipped with BoNT/F7 cluster. The active site was conserved in all strains, identifying a missing 7-aa region upstream of the active site in C. baratii genomes. This analysis could be important to advance our knowledge regarding opportunistic clostridia and better understand their contribution to disease.
spacer
spacer
spacer
spacer
| Genes – National Geographic Society May 20, 2022A gene carries information in the sequence of its nucleotides, just as a sentence carries information in the sequence of its letters. Although most genes are made of DNA, the genes use RNA to make the proteins that are coded for in the DNA. Genes are located on threadlike structures called chromosomes in the cell nucleus. |
| Gene | Definition, Structure, Expression, & Facts | Britannica Genes achieve their effects by directing the synthesis of proteins. In eukaryotes (such as animals, plants, and fungi ), genes are contained within the cell nucleus. The mitochondria (in animals) and the chloroplasts (in plants) also contain small subsets of genes distinct from the genes found in the nucleus. |
| Genes: Function, makeup, Human Genome Project, and research Jul 25, 2022 Gene therapy is a medical technique that uses sections of DNA to treat or prevent a disease or medical disorder. Genes are inserted into a patient’s cells and tissues to treat a disease. |
| Genetics Basics | CDC – Centers for Disease Control and Prevention Genetics research studies how individual genes or groups of genes are involved in health and disease. Understanding genetic factors and genetic disorders is important in learning more about promoting health and preventing (or causing) disease. |
| Genetics – National Institute of General Medical Sciences May 4, 2022 Genetics is the scientific study of genes and heredity—of how certain qualities or traits are passed from parents to offspring as a result of changes in DNA sequence. A gene is a segment of DNA that contains instructions for building one or more molecules that help the body work. DNA is shaped like a corkscrew-twisted ladder, called a double helix. |
spacer

spacer













 What does ‘high-throughput’ mean in sequencing? Alithea Genomics
What does ‘high-throughput’ mean in sequencing? Alithea Genomics